Resources
DYES
Cell Excluded Dyes
Cell Permeant Dyes
Protocol List
Protocol List
Suspension and Handling
How to properly suspend dyes and determine their concentration Protocol-Suspension Concentration Storage
Surface Labeling FAP-Fusions
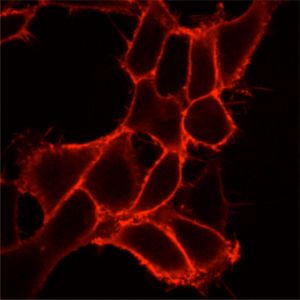
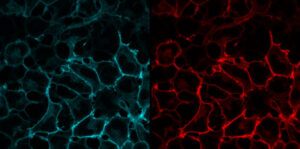
Surface selective labeling is achieved by adding a cell-excluded dye to living cells in a physiological buffer. Labeling is essentially instantaneous, if dye is added at mid-nanomolar concentrations. Protocol-SurfaceLabeling.
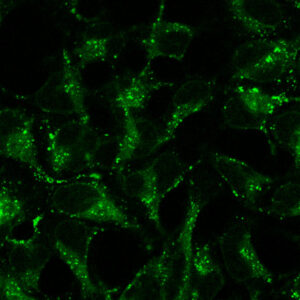
Intraceullar labeling fap-fusions
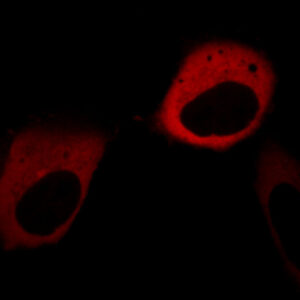
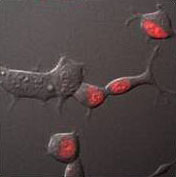
Total protein pool labeling is achieved by adding a cell-permeant dye to living cells in a physiological buffer. After equilibration for a short time, cell-permeant dye penetrates the cell to label intracellular expressed FAP sites. Labeling is rapid (10 min), and dye does not need to be washed out to achieve selective labeling.
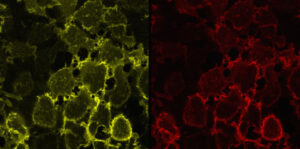
Surface and Intracellular dye labeling combinations
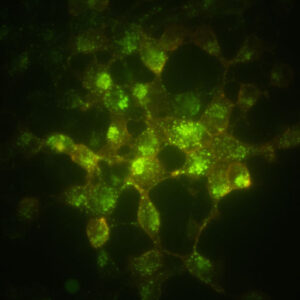
Label distinctive pools of protein in different colors.
Vectors and Plasmid Maps
Vectors
Mammalian Expression
Organelle Targeted FAPs
pcDNA3.1-COX4-COX8-dL5-2XG4S-mCer3
(Mitochondria Addgene 73208)
pcDNA3.1-kappa-myc-dL5-2XG4S-mCer-KDEL
(Endoplasmic Reticulum Addgene 73209)
pcDNA3.1-KozATG-dL5-2XG4S-mCer3
(Cytosolic Addgene 73207)
pcDNA3.1-myc-dL5-2XG4S-mCerSKL
(Peroxisomal Addgene 73210)
pcDNA3.1-NLS-myc-dL5-2XG4S-mCer3
(Nuclear Addgene 73205)
Actin-FAPs
pcDNA3.1-KozATG-dH6.2-2XG4S-actin
Addgene 73263
pcDNA3.1-KozATG-dL5-2XG4S-actin
Addgene 73262
Membrane Proteins
pcDNA3.1-kappa-dL5-2XG4S-TMst
(PDGFR TM anchored surface-expression FAP Addgene 73206)
pcDNA3.1-kappa-HA-dL5-BKa (KCNMA1)
(KCNMA1)
Bacterial Expression
FAP dL5 Bacterial Expression
(Generally, we express from oxidizing strains such as Rosetta B-Gami2)
pET21a-10XHis-GST-HRV-dL5(YW)
(Addgene 74214)
Affibodies and Affi-FAP Constructs
(See Wang, Y et al, Bioconj. Chem. 2015)
pET21a-10XHis-GST-HRV-dHer1cysteineST
(double Her1-affibody Z1907 with cysteine at C-terminus Addgene 73218)
pET21a-10XHis-GST-HRV-dL5-Her1
(FAP-Affibody construct, Her1 Z1907 affibeody Addgene 73215)
pET21a-10XHis-GST-HRV-Her1-dL5-Her1
(Affibody-FAP-Affibody construct, Her1 affibeody Z1907 Addgene 73217)
pET21a-10XHis-GST-HRV-Her1-dL5
(Affibody-FAP construct, Her1 affibody Z1907 Addgene 73216)
Yeast Expression
Plasmid Maps
Transmembrane FAP
pcDNA3.1-kappa-myc-dL5-2XG4S-TMst
pcDNA3.1-kappa-dL5-2XG4S-TMst
Big Potassium Conductance Channel (BKa)
dL5-BKa pcDNA
Nuclear Localized FAP-mCer3 Fusion Protein
pcDNA3-NLS-myc-dL5-2XG4S-mCer
Mitochondrial Localized FAP-mCer3 Fusion Protein
pcDNA3.1-COX4-COX8-dL5-2XG4S-mCer
ER-Localized FAP-mCer3 Fusion Protein
pcDNA3.1-kappa-myc-dL5-2XG4S-mCer-KDEL
Cytoplasmic Localized FAP-Mcer3 Fusion Protein –penetrates nucleus
pcDNA3.1-KozATG-dL5-2XG4S-mCer
________
ApE Plasmid Editor
by M. Wayne Davis
Request for Materials
Carnegie Mellon University
Office: MI-749, Lab: MI-755
4400 Fifth Ave.
Pittsburgh, PA 15213
Phone: (412) 268-9661